Supporting_Scientific_Data
Data Storage and Organization
| Last Chapter - Documentation and Description | Back to the Table of Contents | Next Chapter - Data Sharing |
Chapter Summary
This chapter details strategies and processes related to ensuring that data and other research materials (documentation, code, physical samples, etc) can be found and used as needed.
The chapter is built around the principle that data management must be part of the everyday process of doing science.
| Principles of Good Data Management Practice | “Good Enough” Practices in Data Management |
| Key points for this chapter | |
|---|---|
| 1. | Good data management is built on the foundation of relatively straightforward practices, including those related to the storage and organization of important materials. |
| 2. | Human readability and machine readability are BOTH important properties when storing and organizing research data, they are not interchangeable. |
| 3. | To prevent the loss of data, data should be backed up and kept well organized. Organzing data can involve such rudimentary practices as providing effective names to data-related files. |
| 4. | When possible, it can be helpful to save data in open and/or widely adopted file formats. |
| Supplementary materials for this chapter | |
|---|---|
| Data Index Checklist | This document provides guidance to research teams who need to keep a record of how and where their research data is saved. |
| Fantastic File Names | This exercise guides users through some best practices in naming digital files. Similar principles can also be applied to other names, including the names of variables and column headers. |
5.1. Foundational Practices in Data Management
Picture this:
Our scene opens in an office at an academic institution. Stacks of books and piles of papers crowd every surface. There are multiple filing cabinets, mostly containing printouts of journal articles and boxes containing multiple generations of storage media. Somewhere in here is a three and half inch floppy disc containing an essential piece of scientific data. It is labeled simply “project data” and there are no other copies.
A researcher who is not the main occupant of the office has been tasked with finding the data. They have a deadline and time is running out!
While perhaps amusing, definitely out of date (who has so much paper!), and almost certainly too dramatic, this image of a disorganized academic office helps to illustrate the immense value of relatively simple practices and strategies within Good Research Data Management Practice. Maybe, despite the chaos, the office’s occupant would be able to efficiently find the disk and access the information they need - provided the disk has not degraded and the necessary hardware and software are still available - but anyone else would have a very difficult time.
Loss Prevention
The most immediate benefit of adopting Good Data Management Practice is preventing the loss of data. There are A LOT of ways to lose scientific data. Data can be lost through human error, storage media can fail, software can become corrupted, natural disasters can occur, laptops can be stolen, sweet but clumsy pets can inadvertently knock a computer on the floor rendering it useless1, files can be mislabeled and lost to the abyss of a crowded desktop.
Depending on the circumstance, the consequences of losing data can range from inconvenience to catastrophic. For some forms of research, the cost of re-collecting or reanalyzing data that has been lost is just inefficiency. Re-running a workflow may take some extra time and resources, but the work is still able to proceed. In other cases, re-collection or reanalysis is simply not feasible. The loss of data jeopardizes the entire trajectory of the research effort. Preventing the loss of data through data management involves the implementation of relatively straightforward practices. Though storing and organizing data may seem almost overly simple, such practices go a long way towards ensuring that a research team can find, use, and share their data.
But what about documentation?
This chapter is primarily focused on the implementation of some foundational data management practices. But ensuring maintaining good documentation is perhaps the most foundational practice when managing data. Without sufficient documentation and metadata, even the most exquisitely saved and organized files are less than useful. See the chapter on Documentation and Description for a deep dive into the different forms of documentation and decent recipes for chocolate chip cookies.
Standards and practices
This is true of many aspects of Good Data Management Practice, but when it comes to storing and organizing scientific data, consistency is really key. That said, there is not a single storage strategy or organizational scheme that will work across every area of science. A research team working with a dataset that contains a huge number of electronic health records will have to address a very different set of issues when storing and organizing their data than a team who are recording measurements taken from cells in a spreadsheet.
Elsewhere in this guide, a data standard is defined as just an agreed upon way of organizing, structuring, and/or describing a particular form of research data. In the context of data storage and organization, the term is often invoked in the context of file formats - standardized ways in which information is encoded to be stored in a computer file. This is an extension of what is discussed in the chapter on Documentation on Description, but is also relevant in the context of data storage and organization where the way in which data is structured within a file significantly affects how it can be later accessed and used.
5.2. Storing and Backing Up Files
It is imperative that you properly store and backup your research data.
The statement above is easy to say (or type) but belies some surprising complexity. So before getting into the details of storage and organization, it is important to disentangle a few concepts.
Very, very broadly data storage is just the recording of information into a medium of some kind. Notebooks full of handwritten observations can be considered a form of stored data. DNA and RNA are considered by some to be forms of stored data. But, because of its ubiquity in twenty-first century science, this chapter largely focuses on digital data storage - methods used to store information in an electronic file that can be accessed by a computer.
From radiological images taken from human participants to cell counts taken manually, the contents of a digital file can be just about anything. But that file, ultimately, can be reduced to the most basic unit in computing - a string of values (e.g. 1s and 0s).
Each value is a bit.
A byte is (usually) 8 bits.
But a kilobyte could be 1,012 bytes or 1,000 bytes.
And a megabyte could be 1,048,576 bytes or 1,000,000 bytes.
This is one of those “everybody knows” sort of situations, where different groups may make reasonable assumptions depending on their context. But, due to the absence of agreement or standards, different assumptions lead to increasingly different conclusions2. There’s a lesson here about maintaining good documentation, but for the purposes of Good Data Management Practice, this situation is all mostly useful as another illustration of the complexity around the word “data”.
Wait, what is data again?
This guide defines research data as “the inputs or outputs required to evaluate, reproduce, or build upon the analyses or conclusions of a given research project.” Notice that this definition does not include any reference to megabytes or file formats. This is because, as a guide written by a human for humans, this guide is biased towards human readability - the degree to which data is easily understood and used by people. Research data is part of the research process. Characteristics like file size and format are certainly important properties of the data. But they are, as long as everything is working properly, secondary to how the data is used by researchers to support their findings.
Machine readability, by contrast, is the degree to which a file can be easily understood and used by a computer. Unlike Joe Pantoliano’s Cypher in The Matrix (1999), the average researcher probably cannot look at cascades of binary values and visualize whatever data has been encoded within. But, for a computer, those bytes and the way in which they are organized within a file are absolutely vital. The computer cares much less about the contents.
It is useful to keep these distinctions in mind because any discussion of storing and organizing research data must take BOTH of these perspectives - the needs of the human researchers using the data to do science and the needs of their computational tools to open and use data files. These two perspectives are both important and are, in many ways, complementary. But, addressing one does not necessarily address the other.
Table 5.1. below provides a brief overview of three major categories of data storage media.
Table 5.1. Types of digital data storage media
| Type | Description | Example |
|---|---|---|
| Magnetic | Binary values are encoded on a hard disk or tape as positive or negative polarities. A read/write head then detects the direction of the magnetization and translates it back into usable information. | Hard disc drives (HDD), Floppy disks, Magnetic tape |
| Optical | Binary values are encoded on a reflective surface of a disk. A laser scanner then reads these pits and translates them back into usable information. | Compact Discs (CD), Digital Video Discs (DVD), Blu-ray Discs |
| Solid State | Binary values are encoded as electrical charges in transistors. A controller (processor) then detects these charges and translates them back into usable information. | Solid state drives, Secure Digital (SD) Cards |
Cloud storage, in which digital data is stored remotely rather than on a local storage medium, also ultimately comes down to data stored on one or more of these types of media. While some of the examples listed here are more prone to failure or wear and tear than others, all of them have a rate of failure.
So, to prevent data from being lost, it is really important to maintain backups.
But backup, what is a back-up?
In the context of data management, working data storage refers to the methods and media used to store data that is currently being actively worked on - data that is currently in the process of being transformed, analyzed, and evaluated. The working copy of data stored in a working data storage system is “live” and the goal of working data storage is immediate access and use.
In contrast, a backup data storage refers to the methods and media used to store copies of the working copy that can be used to restore the original if (or when) data loss occurs. The goal of backup data storage is redundancy. The ability to immediately access and use backup copies of a dataset can be helpful, but is not always necessary. Archival storage refers to methods and media that are intended for longer term preservation.
The 3-2-1 Rule
Whenever possible, it is best practice to maintain multiple copies of a set of research data.
A convenient shorthand for this is the 3-2-1 rule3, which states that an individual should maintain a total of three copies of their data, with at least two backups, and one backup that is offsite. This is illustrated in Figure 5.1. Below.
| Figure 5.1. An illustration of the 3-2-1 rule for data storage and backup |
Of course, maintaining multiple backups may not always be feasible.
Throughout science, there are projects that involve the acquisition of extremely large and extremely complex datasets. It is relatively straightforward to store and backup data that is on the scale of megabytes and gigabytes. Strategies and cost effective options narrow considerably when datasets reach sizes counted in terabytes and petabytes. But one backup is better than no backups.
What about sensitive data?
Managing sensitive data - data that must be protected against unauthorized access - is the topic of a whole (forthcoming) chapter, but it is always worth remembering that not every system for storing data provides the same level of security. Think of it this way, a locked filing cabinet provides a much higher degree of security for a paper file than just placing that same file on top of a desk. When considering the security of services for storing data, it is always best to consult with local IT professionals on topics such as encryption, access control, and other measures.
Long term storage
In the not too distant past, data from completed projects was stored on physical media for long-term storage. From magnetic tape and punch cards to boxes of floppy disks and spindles of DVDs to USB flash drives, SD cards, and everything in between, many of these forms of storage are still present in the closets and desk drawers of academic science.
Pretending for a moment that all of these materials are stored under optimal conditions, reducing the risk of disk rot, literal rot, and causes of data corruption, this data is increasingly difficult to access. When preserving scientific data over the long term, there are two interrelated concepts to be aware of:
- Bit rot - Broadly refers to the gradual degradation of the integrity of stored data. This is largely caused by wear and tear on storage media. File fixity is ensuring that a digital file in an archive has remained unchanged at the bit level.
- Obsolescence - In the context of research data management, this refers to an inability to access digital data because the needed hardware or software are no longer available.
Preserving data over the long term is an active process. Let’s put aside physical media temporarily. Hypothetically, let’s say a research team has stored data in a cloud-based system, a box or a drive. That works well enough until that service is retired. Any links that pointed to files in that retired box or drive no longer resolve. This phenomenon, known as link rot, is certainly not unique to scientific research but is especially problematic when such links are necessary to establish data provenance4.
Archival storage generally refers to the storage of data that is not needed day-to-day. There are solutions available for long term data storage that are billed as “set it and forget it”, but it is always worth remembering that access to these services may change over time. This is always true, but especially when archiving data over the long term, remember that simply storing data is not enough. For any kind of data, but for scientific data especially, the stored data is only useful if it can be retrieved and used.
Often research data deposited into a data repository - a platform that facilitates the preservation, organization, and discovery of research data - is said to be archived. This is true to the extent that many repositories have strategies for backing up deposited data for a period of time. But it is also true that the data deposited into a repository is not ALL of the data associated with a given research project. Perhaps only the processed “final” dataset is uploaded, while storage of “raw” and “intermediate” forms are left to the research team.
Tool Highlight: Data Index Checklist
Over the last few years, tools like Google Dataset Search and the NLM dataset catalog have emerged to help researchers find data shared by others. These tools are useful, but limited by the fact that only a portion of the data related to a project may ultimately be shared.
This checklist is intended for groups who need to preserve the rest of their data themselves.
5.3. File Formats and Naming Things
As we promised in the introduction, this guide does not advocate for the use of one software tool over another. However, that does not preclude a word of caution about the use of an overwhelmingly popular spreadsheet tool. More to the point, the ubiquity of Excel spreadsheets in and outside of scientific research provides a nice entrypoint to the exciting world of file formats.
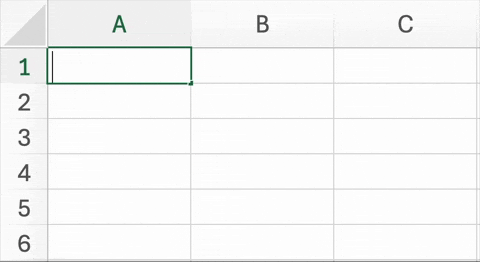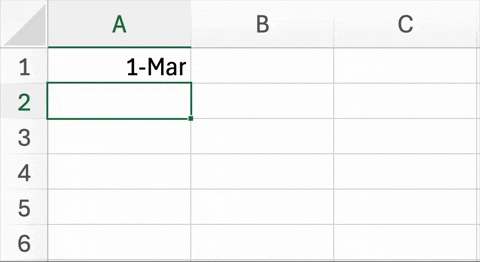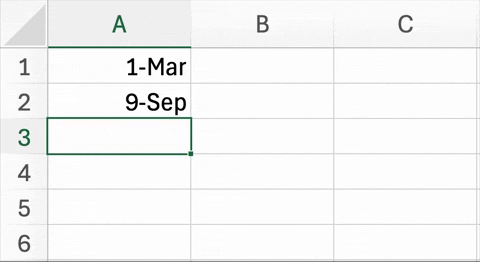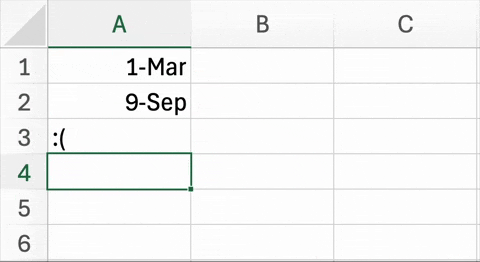
Figure 5.2. below is an example of a well known and widespread problem5. In trying to simplify data entry, this Excel spreadsheet has changed the names of two genes to dates. Unfortunately, it is not just the display of this information that is being changed, but the underlying content of the cells. This is not a critique of Excel (or geneticists) per se. In recent versions of Excel, there are ways to disable automatic date conversion. Also, the HUGO Gene Nomenclature Committee (HGNC) recently recommended that the names of these particular genes be changed to MARCHF1 and SEPTIN1 to avoid this problem6.
 |
|---|
| Figure 5.2. An example of Excel automatically changing two genes into dates. The abbreviated names for the Membrane Associated Ring-CH-Type Finger 1 (MARCH1) and Septin-9 (SEPT9) genes are changed March 1st and September 9th, respectively. |
Choosing a file format (when possible)
As already stated, file formats are standard ways that information is encoded for storage in a computer file. But, for the purposes of research data management, there are two axes that are important.
- Openness - Many file formats are proprietary (or closed), meaning that they encode data in a manner that is secret or restricted in some way. Proprietary file formats typically can only be opened and used with specific software tools. In contrast, open (non-proprietary) file formats are unrestricted and free to use.
- Lossiness - When information is encoded into a file, it is often compressed in some way. “Lossy” compression involves the removal of information (and thus decreases the fidelity of the information) while “lossless” compression does not.
Choosing the right file format, when there even is a choice, is by no means an uncomplicated decision.
Here’s an example: If you are old enough to have used physical media to listen to music, you may also remember the process of converting the highlights of your CD collection into digital files. Converting to .mp3 - a lossy file format - was (and remains) very popular because of the relatively small size and ubiquitous support for playing the resultant files. However, in doing so, the fidelity of the audio was compromised. Audiophiles would perhaps convert to FLAC format - a lossless file format - but those files were substantially larger and able to be played by a fewer number of programs and devices.
Here’s another example: Around the time you were converting your CDs into digital files, you may have been writing documents and managing spreadsheets in ClarisWorks (or AppleWorks), which saved files in .cwk format. Hopefully, you were not working on anything too important, because this program is no longer supported. Converting these files into something usable nowadays is, at best, a tricky process. So much so that a common workaround is to use a MacOS emulator.
Balancing openness, lossiness, storage costs, and other considerations in choosing the right file format for digital data can feel a bit like trying to do advanced calculus. It also might not be something you have control over. If you are collecting or analyzing data with a particular instrument (microscope, MRI scanner, sequencer, etc), you may not have control over the formats of the files it requires or outputs. A non-comprehensive list of formats that are open and/or recommended for preservation purposes is presented in Table 5.2, but best practice on this issue can be summed up thusly:
When saving files, try to save in file formats that are non-proprietary and/or commonly adopted. That will prevent headaches when trying to use those files in the future.
Table 5.2. A non-exhaustive list of file formats
| Modality | Open Formats | Formats for Preservation7 |
|---|---|---|
| Text | Plain text (.txt), Markdown (.md), LaTeX (.tex), Portable Document Format (.pdf), Office Open XML (.docx), OpenDocument (.odf) | Electronic Publication (.epub), Portable Document Format (.pdf), XML-based document formats (e.g. .docx, .odf, etc) |
| Images | Graphical Interchange Format (.gif), JPEG (.jpeg), Portable Network Graphics (.png) | Tag Image File Format (.tiff), JPEG-2000 (.jp2), Portable Network Graphics (.png) |
| Video | Matroska (.mkv) | Matroska (.mkv), MPEG-4 (.mp4) |
| Audio | MP3 (.mp3), Free Lossless Audio Codec (.flac) | Waveform Audio File Format (.wave) |
| Spreadsheets and databases | Tab separated values (.tsv), Comma separated values (.csv), OpenDocument Spreadsheet (.ods), OpenDocument Database (.odb), SQLite (e.g. .db, .sqlite, etc) | Platform independent, character-based formats (.tsv, .csv, .db, .sqlite, etc) or proprietary formats that are de facto standards for a profession and/or supported by multiple tools (e.g. Excel) |
Naming Things
Storing and backing up data properly is vital, but maintaining multiple copies of a dataset is only valuable if the files can actually be found. We promise this is not actually a screenshot of anyone’s desktop, but Figure 5.2. demonstrates the necessity of keeping data-related materials well organized. This is almost as bad as the anecdote that leads off this chapter. If a researcher were asked to pick the most recent file from this display, it would be quite the challenge.
What is the difference between data_v2? and Data2?
What is the difference between data_v2 final copy and data_v2 final?
| Figure 5.2. An illustration of the necessity of maintaining a good file naming scheme through the depiction of a lack of good file naming. The author of this guide promises that this is not the contents of his desktop |
It may seem almost too rudimentary, but providing good names to things (files, variables, etc) is actually one of the cornerstones of good data management. Compare the following two files. Hopefully you can tell a lot more about one than the other just by looking at their names.
` data.csv `
This provides a limited amount of information. A user knows that this file is data, but for which research project? When was it was it modified and by whom? How does this file differ from other data-related files?
` Borghi_rdm-project_participant-data_2024-12-31.csv `
This provides some additional information. There is a researcher listed (this guide’s author), a project name (very creative), some description of the file’s contents, and a date that’s useful for tracking versions. But for this kind of file name to be truely useful, it needs to be used consistantly.
A file naming convention is a consistent way of naming files in a way that provides information about the contents of the file and how it relates to other files. A research group’s conventions related to naming files should be documented as part of their standard operating procedures. Table 5.3 provides some practical tips for naming files.
Table 5.3. Practical tips when naming files
| Tip | Description |
|---|---|
| Determine what information would be useful | When putting together a file naming convention, first determine what information will make the name useful. Probably it will be more than one piece of information (project name, investigator name, identifiers, dates, version, etc). |
| Establish a consistent order for information within your file names. | Keeping the various pieces of information in a filename in the same order will help make sorting and searching easier. |
| Establish a consistent way of identifying different versions | There are lots of ways of marking different versions of the same file. If using version numbers, make sure you’re using leading zeros (e.g. v01 instead of v1), so sorting and searching will be easier. If you’re using dates, which we typically recommend, consider using YYYY MM DD8 for the same purpose. |
| Keep file names (relatively) short and do not use spaces and special characters | There is a hard limit on the number of characters in a file path (256 in Windows, for example). This may seem like a lot, but does demonstrate the need to keep file names as pithy as possible while still being informative. Whenever possible, file names should only use only Latin alphanumeric characters (a-z, 0-9), dashes, and hyphens. This is mostly because of issues with the command line. Spaces can cause file names to be cut off and other characters may have specific functions (and different functions for different operating systems) that would cause issues if included in the name of a file. |
| Document your convention | Record the naming convention you come up with, as well as any abbreviations or code. This information can be articulated as a standard operating procedure (SOP) Information about the contents of data files (e.g. variable names, units, etc) and what has changed between versions should also be thoroughly documented (more on that in the next section). |
File names may, of course, be part of a broader data standard. But researchers may find themselves in a position where they have to come up with a convention more or less from scratch. To facilitate this process, see the Fantastic File Names exercise.
Tool Highlight: Fantastic File Names
Sometimes file names are described as part of a broader data standard or are automatically determined by the software being used alongside the data. This exercise guides users through some best practices in naming digital files when neither of these situations apply. Similar principles can also be applied to other names, including the names of variables and column headers.
Versioning Files and Version Control
When working on a research project, it is quite likely that keeping track of a research workflow will necessitate some degree of version control. This term refers to systems that are responsible for managing changes in software and code, documents, and other sets of information.
Version control is probably (especially here in the Bay Area) most commonly invoked in a software development context, where a large number of people may be working on the same materials simultaneously. Tools like Github, Bitbucket, and SourceForge fulfill a large number of functions, but their primary purpose is to facilitate version control.
This type of version control can be used for some types of scientific data. Indeed, there is quite a bit of scientific data hosted on platforms such as Github. However, this is not feasible for many types of data or many research groups. The data may be too big or two sensitive to be uploaded to a public repository while the research process is active. A research group may have tremendous expertise in many areas, but may not have the time or need to learn the ins and outs of “the stupid content tracker9”. In these cases, versioning is still important but may involve a more manual set of strategies and behaviors.
These include:
- Including version-related information in filenames. This can be as simple as a sequence of numbers (e.g. v01, v02, etc). But we would recommend considering the use of dates (e.g. 2024-01-29, 2024-01-31) to make sorting and searching easier. The difference between file versions should be well documented, in a change log or lab notebook.
- The use of tools that have versioning built in. We have already covered the need to be considerate about what tools you use to save your data. Some tools make it straightforward to identify when changes were made and by whom. Others… less so.
- A combination of the above. This would be ideal, but is not always possible.
But remember, the most important thing is to be consistent.
File Folders and Directories
Let’s say a research team has their files properly saved and backed up. They also have a good system for naming things and demarcating different versions. But what if somebody else needs to look at what they have done? Would this individual know that every file is in the downloads folder of the PI’s computer? How loud of a sigh would the research team hear if somebody else tried to access project-related files without team members around to help them?
Another cornerstone of good data management is maintaining an easily navigable file directory structure. For individuals who do the majority of their computing on a tablet or other mobile device, a directory (e.g. a file folder) is a location for storing files in a computer system. Directories can be organized in a hierarchical manner, with nested subdirectories. Figure 5.3 provides a few examples of how files can be organized.
There are some standardized ways to organize research project folders, including Project Tier, Cookie Cutter Data Science and Gin-Tonic. But the precise organization of a set of files will depend on the needs of the research team. Some data standards impose an organizational structure. But in settings where there are no widely adopted standards, consistency - as always - is key. Project folders for different projects and maintained by different members of the research team should have the same basic structure. One member of the research team should be able to open any project folder and have a general sense of how to navigate it.
Table 5.3. Example directory structures
| Illustrated Example | Description |
|---|---|
| A relatively flat file hierarchy. In this example, a relatively small number of the same type of files need to be kept organized. Specifically, within a project folder, there are three versions of one file (helpfully labeled as project data) and a notes document that contains information about the other files. Because of the small number of files, a more complex structure is not needed. In practice, a situation like this is unlikely. | |
| A more complex file hierarchy. In this example, a (relatively) large number of different types of files need to be kept organized. To facilitate this, the project folder contains subfolders - one for data, one for documentation, and another for scripts. The subfolders may contain subfolders of their own, for example the data folder contains folders for raw, processed, and finalized data. A readme file is maintained at the top of this file structure, to help make everything more navigable. This file, often a plain text file, contains information about the structure of the project file. Reading it should help someone opening the file for the first time understand where to look for the files they need. In practice, it is likely that project files will be even more complex than this example. |
“Good Enough”
There will be times when an individual researcher - especially those working in academic settings - will simply not be in position to dictate how everyone in their group is saving and organizing their data. This is why there is a whole chapter devoted to “Good Enough Practices” in data management. But, returning to the example that began this chapter, an ideal to shoot dor is something closer to the following:
Our scene closes in an office at an academic institution. There are stacks of books and papers on every surface. The multiple filing cabinets have data stored for the last several years of experiments on a stable form of storage media, with additional copies backed up to the cloud. There is an index on the lab’s internal server saying how data can be located. Some of the data has also been stored in data repositories, meaning it can be accessed and reused without much intervention.
There is now even more room for printouts of journal articles.
(Please use a citation management tool like Zotero to address this last sentence).
Definitions of Key Terms
3-2-1 Rule - A good rule of thumb to ensure that data is not lost. Stipulates that 3 copies of data should be maintained (1 working copy, 2 backups) and that at least one of the backups should be in saved different location.
Archival Storage - Refers to methods and media used to store data that is not needed day-to-day. The goal of archival storage is long-term preservation.
Backup Storage - Refers to methods and media used to store copies of data that can be used to restore the original if (or when) data loss occurs. The goal of backup data storage is redundancy.
Bit Rot - Broadly refers to the gradual degradation of the integrity of stored data. This is largely caused by wear and tear on storage media.
Cloud Storage - A system in which digital data is stored remotely rather than on a local storage medium.
Data Repository - A platform that facilitates the preservation, organization, and discovery of research data.
Data Storage - The recording of information into a medium of some kind.
Directory (e.g. File Folder) - A structure that contains computer files and possibly other directories.
File Naming Convention - A consistent way of naming files in a way that provides information about the contents of the file and how it relates to other files.
File Format -Standardized ways in which information is encoded to be stored in a computer file. Think .XLSX, .CSV, etc.
File fixity - The process of ensuring that a digital file in an archive has remained unchanged at the bit level.
Lossiness - The degree to which information is lost when information is encoded into a particular file type. “Lossy” compression involves the removal of information (and thus decreases the fidelity of the information) while “lossless” compression does not.
Obsolescence - Refers to an inability to access digital data because the needed hardware or software are no longer available.
Openness - The degree to which the way a file type encodes information in a manner that is secret or restricted. Proprietary file formats typically can only be opened and used with specific software tools. In contrast, open (non-proprietary) file formats are unrestricted and free to use.
Version Control - Systems that are responsible for managing changes in software and code, documents, and other sets of information. Can refer to tools like Git but also to more manual forms of keeping track of versions, such as using dates or version numbers in file names.
Working Data Storage Refers to the methods and media used to store data that is currently being actively worked on - data that is currently in the process of being transformed, analyzed, and evaluated. The goal of working data storage is immediate access and use.
A Bonus GIF
Among the many reasons why Excel is not always the best choice for storing data is that a lot of what it is doing at any given time immediately apparent to the user. What works for doing some quick calculations and setting up some fun macros does not necessarily work for maintaining research data provenance.
 |
|---|
| Figure 5.3. Tetris running in an Excel spreadsheet.. There are many different versions of this available online, but finding a version that works with a specific version of Excel can be challenging. Another reason to consider storing data in more open formats (e.g. CSV) is future proofing. Presumably if a researcher is saving some data, they will want to be able to open and use it in the future. |
Further Reading
Cookie Cutter Data Science - A flexible standard for organizing and sharing data science-related project
Gin Tonic - A simple adult beverage for those who partake (1 part gin, 2 parts tonic water, poured over lots of ice). Also a proposed standard for keeping research-related files organized in a common file structure.
TIER Protocol - A specification for organzing the materials needed to reproduce the work underlying a research-related manuscript.
Footnotes
-
This happened to this chapter’s author ahead of his qualifying exams. Another hard drive failed right before his PhD defense. ↩
-
For more on how this happened and an explanation of terms like kibibit and mebibit, see Barrow, B. (1997). A Lesson in Megabytes. IEEE Standards Bearer, 5. Megabyte is also the main antagonist of the extremely 90’s animated series Reboot. That doesn’t really matter for data management purposes. Just checking that folks are reading the footnotes. ↩
-
Now ubiquitous, this rule was first described by Peter Krogh in his 2005 book “The DAM Book: Digital Asset Management for Photographers”. ↩
-
For more on this, see: Wren, J. D. (2004). 404 not found: The stability and persistence of URLs published in MEDLINE. Bioinformatics, 20(5), 668–672. https://doi.org/10.1093/bioinformatics/btg465 ↩
-
Zeeberg, B. R., Riss, J., Kane, D. W., Bussey, K. J., Uchio, E., Linehan, W. M., Barrett, J. C., & Weinstein, J. N. (2004). Mistaken Identifiers: Gene name errors can be introduced inadvertently when using Excel in bioinformatics. BMC Bioinformatics, 5(1), 80. https://doi.org/10.1186/1471-2105-5-80 ↩
-
Bruford, E. A., Braschi, B., Denny, P., Jones, T. E. M., Seal, R. L., & Tweedie, S. (2020). Guidelines for human gene nomenclature. Nature Genetics, 52(8), 754–758. https://doi.org/10.1038/s41588-020-0669-3 ↩
-
These recommendations are adopted from the Library of Congress’s Recommended Formats Statement - https://www.loc.gov/preservation/resources/rfs/ ↩
-
This format is part of a data standard for the representation of date and time-related information (ISO 8601). Using this format not only makes sorting and searching easier, but sidesteps a quirk of how dates are expressed throughout the world . Is February 1st commonly represented by 02/01 or 01/02? It depends on where you live! ↩
-
This nickname for Git, the system that underlies Github and many other version control tools, comes straight from the user manual. It can actually mean anything from “Global Information Tracker” to much ruder acronyms, depending on your mood. ↩